Autoencoder for Ultrasound Images¶
In this notebook, we demonstrate how to use the TAESD tiny autoencoder model for ultrasound images using the zea toolbox. We’ll encode and decode images, visualize the results, and try a simple experiment: interpolating (traveling) in the latent space between two images and animating the result. Note that this model was trained purely on image data, and not finetuned on ultrasound data.
[1]:
%%capture
%pip install zea
[2]:
import os
os.environ["KERAS_BACKEND"] = "tensorflow"
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
[3]:
import matplotlib.pyplot as plt
import numpy as np
from keras import ops
from matplotlib import animation
from IPython.display import HTML
import zea
from zea import init_device
from zea.backend.tensorflow.dataloader import make_dataloader
from zea.models.taesd import TinyAutoencoder
from zea.visualize import plot_image_grid, set_mpl_style
zea: Using backend 'tensorflow'
We will work with the GPU if available, and initialize using init_device to pick the best available device. Also, (optionally), we will set the matplotlib style for plotting.
[4]:
init_device(verbose=False)
set_mpl_style()
Load data¶
We load a small batch from the CAMUS validation dataset hosted on Hugging Face Hub.
[5]:
n_imgs = 4
val_dataset = make_dataloader(
"hf://zeahub/camus-sample/val",
key="data/image",
batch_size=n_imgs,
shuffle=True,
image_size=[256, 256],
resize_type="resize",
image_range=[-60, 0],
normalization_range=[-1, 1],
seed=42,
)
batch = next(iter(val_dataset))
zea: Using pregenerated dataset info file: /home/devcontainer15/.cache/zea/huggingface/datasets/datasets--zeahub--camus-sample/snapshots/617cf91a1267b5ffbcfafe9bebf0813c7cee8493/val/dataset_info.yaml ...
zea: ...for reading file paths in /home/devcontainer15/.cache/zea/huggingface/datasets/datasets--zeahub--camus-sample/snapshots/617cf91a1267b5ffbcfafe9bebf0813c7cee8493/val
zea: Dataset was validated on July 21, 2025
zea: Remove /home/devcontainer15/.cache/zea/huggingface/datasets/datasets--zeahub--camus-sample/snapshots/617cf91a1267b5ffbcfafe9bebf0813c7cee8493/val/validated.flag if you want to redo validation.
zea: WARNING H5Generator: Not all files have the same shape. This can lead to issues when resizing images later....
zea: H5Generator: Shuffled data.
I0000 00:00:1753082381.837601 1615057 gpu_device.cc:2019] Created device /job:localhost/replica:0/task:0/device:GPU:0 with 9804 MB memory: -> device: 0, name: NVIDIA GeForce RTX 2080 Ti, pci bus id: 0000:1e:00.0, compute capability: 7.5
zea: H5Generator: Shuffled data.
Load TAESD Model¶
We use the built-in TAESD model from zea.
[6]:
model = TinyAutoencoder.from_preset("taesdxl")
Encode and Decode¶
We encode and decode the images using the autoencoder. TAESD expects grayscale or RGB, so we keep the input as is.
[7]:
recon = model(batch)
mse = ops.convert_to_numpy(ops.mean((recon - batch) ** 2))
print("MSE (reconstruction):", mse)
I0000 00:00:1753082385.483177 1615323 cuda_dnn.cc:529] Loaded cuDNN version 90300
MSE (reconstruction): 0.00917655
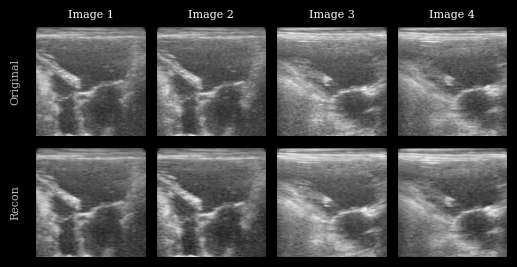
Visualization¶
We plot the original images and reconstructions for comparison.
[8]:
imgs = [zea.display.to_8bit(batch[i], (-1, 1), pillow=False) for i in range(n_imgs)] + [
zea.display.to_8bit(recon[i], (-1, 1), pillow=False) for i in range(n_imgs)
]
titles = [f"Image {i + 1}" for i in range(n_imgs)]
fig, _ = plot_image_grid(
imgs,
ncols=n_imgs,
remove_axis=False,
figsize=(n_imgs * 2, 3),
)
for i, ax in enumerate(fig.axes[:n_imgs]):
ax.set_title(titles[i], fontsize=8)
fig.axes[0].set_ylabel("Original", fontsize=8)
fig.axes[n_imgs].set_ylabel("Recon", fontsize=8)
plt.show()
Latent Space Interpolation¶
Let’s pick two images, encode them, and interpolate between their latent representations. We’ll decode each interpolated latent and animate the result to visualize a smooth transition between two ultrasound images. Again, note that this model was trained on natural images only, and improved results are expected with a finetuned model on ultrasound data (to be added!).
[9]:
# Pick two images to interpolate between
img_a = batch[0][None, ...]
img_b = batch[2][None, ...]
latent_a = model.encode(img_a)
latent_b = model.encode(img_b)
n_steps = 12
alphas = np.linspace(0, 1, n_steps)
frames = []
for alpha in alphas:
latent_interp = (1 - alpha) * latent_a + alpha * latent_b
recon_interp = model.decode(latent_interp)
frames.append(zea.display.to_8bit(recon_interp[0], (-1, 1), pillow=False))
fig, ax = plt.subplots(figsize=(5, 4), dpi=50)
im = ax.imshow(frames[0], cmap="gray", vmin=0, vmax=255)
ax.axis("off")
def animate(i):
im.set_data(frames[i])
return [im]
interval = 40
ani = animation.FuncAnimation(fig, animate, frames=len(frames), interval=interval, blit=True)
plt.close(fig)
HTML(ani.to_jshtml(fps=1000 // interval, embed_frames=True, default_mode="reflect"))
[9]:
You should see a smooth morphing between two ultrasound images, demonstrating the structure of the TAESD latent space.

